What’s NEW with the PSTP?
Two new initiatives will uniquely prepare students for careers in the biotechnology and pharmaceutical industries.
- An NIH-supported laboratory course in the biophysical characterization of therapeutic mAbs, antibody-drug conjugates and other platforms. These protein therapeutics (‘biologics’) have become the largest part of the pharmaceutical pipeline, but few academic programs train students in this area.
- An industry mentoring program with an expanded industry internship. Students will have access to industry scientists for career advice and scientific collaboration. Many will have the option of doing 10-week internships at companies.
Attention Current PSTP Trainees and Alumni!
We are continually working to improve our program and are actively soliciting feedback from our trainees, past and present. Here’s a link to the PSTP Program Evaluation.
The pre-doctoral Pharmacological Sciences Training Program (PSTP) at the University of Washington (UW) is a cross-disciplinary program that represents a merger of research training opportunities in the Schools of Medicine (Dept. Pharmacology) and Pharmacy (Depts. Medicinal Chemistry and Pharmaceutics). PSTP provides traineeships for 12 graduate students working towards their PhD in the broad areas of “Drug Action, Metabolism and Kinetics,” but following more specific tracks that emphasize training in:
- cellular and molecular pharmacology;
- structure and drug/vaccine design;
- drug metabolism;
- pharmacokinetics/dynamics and drug transport.

Med Chem Professor Bill Atkins visits with Dr. Brenda Kelly during a recruiting trip to Gustavus Adolphus College in Minnesota. Kelly, a former Training Grant recipient, is Associate Professor and Chair of the Department of Biology and Chemistry at the College.
The program provides essential in-depth training in several inter-related disciplines central to current and future research related to the discovery, validation, and development of new drug targets and new chemical entities that will improve global health.
Research in molecular mechanisms of drug action, metabolism, and pharmacokinetics constitute core activities of training grant faculty in all three departments. Faculty in pharmacology at the UW have long been recognized for ground-breaking research in signal transduction, ion channel structure and function, neuropharmacology, drug addiction, stem cell biology, behavior and memory; while Medicinal Chemistry and Pharmaceutics are unique in providing integrated, advanced training in the disciplines of drug disposition (metabolism and transport), pharmacokinetics, and pharmacodynamics.
The PhD degree programs of the three departments have many operational similarities with some differences:
- Each department recruits graduate students and makes admission decisions individually, based on academic record, letters of recommendation, research experience, and GRE scores.
- TOEFL scores are required of international students.
- Each department has its own required courses, research rotations, and seminars in the first year, and responsibility for monitoring progress of its graduate students.
Students choose a research advisor at the end of the first year. Research rotations in the first year may be done with faculty in any department, regardless of the graduate program of entry to the University. Students may transfer among graduate programs of departments by standard procedures of the Graduate School of the University.
Entry into the Pharmacological Sciences Training Grant Program (NIGMS National Research Service Award in Pharmacological Sciences) is by competitive application of eligible students (U.S. citizens) during their first three years in graduate school (**link to application form**). We intend to select the most highly qualified students into the Program, while maintaining appropriate breadth across the three core disciplines. Appointment to the PSTP provides opportunities to participate in program-specific Pharmacology courses, local scientific retreats, career seminars, and reseach-relevant biostatistics courses.

The Society for the Advancement of Chicanos and Native Americans in Science, an award-winning organization whose mission is to “foster the success of scientists from diverse backgrounds to attain advanced degrees, careers and positions of leadership in science,” is well represented by past Vice President, Natalie Garcia. Our UW group was named Chapter of the Year in 2010-2014. Visit the SACNAS website for more information.
Students apply to the Program in early Spring Quarter, with successful applicants appointed on July 1 of each year. Students are supported for a maximum of 24 months. Subsequent support, if needed to complete the PhD degree, comes from research grant sources of the major professor.
The Pharmacological Sciences Training Grant is committed to recruiting academically underrepresented graduate students to the PSTP. We value and honor diverse experiences and perspectives, strive to create welcoming and respectful learning environments, and promote access and opportunity for all of our students.
Please see the many resources on campus listed below that are available to help students of all backgrounds and circumstances. Additional information can be obtained by contacting the Program Director, Bill Atkins, PhD.
Disability Services Office
http://www.washington.edu/admin/dso/
Graduate Opportunities and Minority Achievement Program (GO-MAP)
https://grad.uw.edu/diversity/go-map/
Society for the Advancement of Chicanos and Native Americans in Science (SACNAS)
http://students.washington.edu/sacnas/
Native Life and Tribal Relations
http://www.washington.edu/diversity/tribal-relations/
Alliance for Students with Disabilities in Science, Technology, Engineering, and Mathematics
http://www.washington.edu/doit/
Participating Faculty
Department of Medicinal Chemistry
Alonge, Kimberly M., Ph.D. Characterization of chondroitin and heparan sulfate glycosaminoglycan sulfation patterning and their role in neurological processes including neurodevelopment, central control of metabolism, and Alzheimer’s disease; method development for mass spectrometry imaging of glycans.
Atkins, William M., Ph.D. Structure-function mechanism of glutathione S-transferases and cytochrome P450; protein engineering of supramolecular aggregates.
Bhardwaj, Gaurav, Ph.D. Design and development of peptide-based therapeutics for antibiotic, antiviral and anti-cancer applications.
Guttman, Miklos, Ph.D. Characterizing the structures and biophysical properties of complex antigen-antibody interactions; development of methods for glycosylation analysis.
Lee, Kelly K., Ph.D. Biophysical studies of conformational dynamics in viruses. Influenza Hemagglutinin: Structure, Dynamics and Cooperativity During Fusion.
Nath, Abhinav, Ph.D. Conformational dynamics governing enzyme catalysis and protein aggregation; single-molecule fluorescence spectroscopy; computational methods.
Rettie, Allan E., Ph.D. Drug metabolism: Structure-function relationships and pharmacogenetics of P450.
Totah, Rheem, Ph.D. Extrahepatic cytochrome P450 enzymes involved in drug-induced tissue-specific toxicity.
Xu, Libin, Ph.D. Mechanism of lipid oxidation and metabolism and their roles in human diseases; cholesterol biosynthesis disorders; metabolomics and lipidomics using ion mobility-mass spectrometry.
Department of Pharmaceutics
Ho, Rodney J.Y., Ph.D. Enhancement of anti-cancer and anti-infective therapy through targeted drug delivery to lymphoidal tissue; targeted drug delivery to the brain and mechanisms of drug resistance.
Hu, Shiu-Lok, Ph.D. Mechanisms of host-viral pathogen interactions and the development of effective HIV-1 vaccines; animal models for HIV/AIDS research.
Isoherranen, Nina, Ph.D. Regulation of all-trans-retinoic acid homeostasis and involvement of CYP26 activity; mechanisms of xenobiotic-induced teratogenicity; metabolically-based drug-drug interactions.
Kelly, Edward J., Ph.D. Function of CYP4 enzymes in normal and disease states; Generation and characterization of transgenic/knockout mice as preclinical models of human gene function; Development of hepatocytes from human embryonic cells.
Klatt, Nichole, Ph.D. Understanding mechanisms underlying mucosal dysfunction and development of the latent viral reservoir during HIV infection, and developing novel therapeutic interventions to treat HIV disease pathogenesis and prevent HIV acquisition.
Lin, Yvonne, Ph.D. Regulation of drug metabolizing enzymes in children; effect of obesity and diabetes; metabolomics.
Mao, Qingcheng, Ph.D. Drug disposition mediated by BCRP; maternal-fetal drug transport in the placenta and into breast milk; regulation of drug transporters.
Prasad, Bhagwat, Ph.D. Characterization of inter-individual variability in drug disposition in children; pediatric physiologically based pharmacokinetic modeling; Quantitative mass spectrometry proteomics; Drug disposition and toxicity of ester, amide, and lactone containing drugs.
Thummel, Kenneth, Ph.D. Mechanisms of inter-individual variability in drug clearance; role of DMEs in adverse drug responses; pharmacogenetics – molecular mechanisms, clinical translation and public policy.
Unadkat, Jashvant D., Ph.D. Mechanisms of transport and metabolism of drugs to treat HIV, AIDS-associated infections and cancer; drug disposition, DDIs and adverse drug response; cell efflux transporters and disease risk.
Wang, Joanne, Ph.D. Novel drug and neurotransmitter transporters in the CNS, and association with drug efficacy and disease risk; impact of drug transport on oral bioavailability and renal clearance processes.
Department of Pharmacology
Catterall, William A., Ph.D. Molecular basis of electrical excitability; regulation of excitability of nerve and muscle cells; mechanism of actions of drugs and toxins that affect electrical excitability; molecular biology of ion channels; molecular mechanisms in epilepsy.
Chavkin, Charles, Ph.D. Molecular signal transduction mechanisms mediating the addictive properties of drugs of abuse: morphine and cocaine.
Cirulli, Vincenzo, Ph.D. Function of highly specialized proteins that regulate cell-cell and cell-matrix interactions in the pancreas.
Cook, David, Ph.D. Molecular mechanisms of Alzheimer’s disease.
Kalume, Franck, Ph.D. Investigating the pathophysiological mechanisms of a form of epilepsy called Dravet Syndrome, and the mechanism that allows the ketogenic high-fat diet to suppress seizures.
Ong, Shao-En, Ph.D. Quantitative proteomics approaches to investigate the role of protein complexes and their interactions in cellular signaling, gene expression and transcriptional regulation.
Phillips, Paul, Ph.D. Phasic dopamine release in motivated behavior, decision making, and mental illness.
Sancak, Yasemin, Ph.D. Regulation of mitochondrial functions by signaling, regulation of cellular signaling and transcription by mitochondria.
Scott, John, Ph.D. Specificity of signal transduction events that are controlled by anchoring proteins, which facilitate rapid signal transduction by optimally positioning protein kinases and phosphatases in the vicinity of their activating signals and close to their substrates.
Shechner, Dave, Ph.D. Noncoding RNAs and the molecular basis of mammalian nuclear architecture. Assembly and function of subnuclear structures throughout cellular differentiation and disease. Mammalian RNAs and RNA-binding proteins as therapeutic targets. Novel RNA-based and RNA-focused technology development.
Stella, Nephi, Ph.D. Cells and receptors controlling neuroinflammation.
Wang, Edith H., Ph.D. Molecular mechanisms of gene transcription in cell proliferation, differentiation and human disease.
Yadav, Smita, Ph.D. Elucidating the role of kinase signaling in neuronal development and disease using chemical-genetics, proteomics and stem cell techniques.
Zheng, Ning, Ph.D. Structural biology of protein ubiquitination and protein degradation.
Zweifel, Larry, Ph.D. Genetic dissection of neural circuit function and behavior. Understanding the mechanisms of phasic dopamine-dependent modulation of reward and punishment and the role dopamine in generalized fear and anxiety.
PSTP Alumni
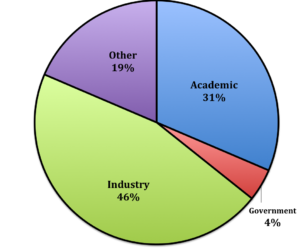
The success of the PSTG over the past 15 years is clear, based on standard metrics, as summarized below. In general, about half of our trainees are now in careers in the pharmaceutical or biotechnology industries. In some cases they accept an academic postdoctoral position before entering the industry sector, but in many cases they go directly into industry careers immediately after earning their Ph.D. Among these graduates, former trainees of the program hold executive and high-level management positions at major companies including Amgen, Merck, Pfizer, Seattle Genetics, Genentech, Lily, and many others. We also strive to place interested students in academic positions. These trainees are nearly always successful in getting postdoctoral positions in excellent labs at top-notch universities, which is the first step toward getting a faculty position. Fewer students choose to go to the FDA or other government labs and we support those students in their job searches. Our trainees complete their degrees in approximately 5.6 years after entering their respective departmental graduate programs. Importantly, the overall graduation rate of trainees is nearly 100%. We work hard to ensure that students successfully complete the program once they are admitted.
Outcomes for our PSTP Alumni:
- Number of trainees over the past fifteen years: 70
- Percent graduated: 90%
- Average time to degree completion: 5.89 years
- Career types our graduates are obtaining after earning their PhD: